Our Work ALIGNED is Accepted by ICLR’26
It’s really delighted to have the opportunity to present our work at a high-impact conference in AI & ML, the International Conference on Learning Representations (ICLR)! This is my first time to publish a peer-reviewed academic output as first author.
Despite negative review feedbacks and accidents occurred in the ICLR’26 review process, we finally made a good enough with max. score 10, really despaired when seeing this rebuttal response, which led to a (marginal?) accept. While acknowledging that our work has many limitations (and indeed some of that implies a low score, from the deep learning community), this experience has also shown me the difficulty of filling the cognition gap between (biological) knowledge-driven perspectives and the deep learning community.
with max. score 10, really despaired when seeing this rebuttal response, which led to a (marginal?) accept. While acknowledging that our work has many limitations (and indeed some of that implies a low score, from the deep learning community), this experience has also shown me the difficulty of filling the cognition gap between (biological) knowledge-driven perspectives and the deep learning community.
ALIGNED follows my line of attempt in integrating biological knowledge bases with transcriptomic data-driven learning, which begins at iGEM in early 2024 and has largely guided my early research explorations. The framework updated for 4 major versions, from a prototype at the iGEM competition to an innovative idea that can be accepted by ICLR. Here, I would like to thank for all supports I’ve received till now, on this ongoing journey.
ALIGNED: Adaptive aLignment of Inconsistent Genetic kNowledgE and Data
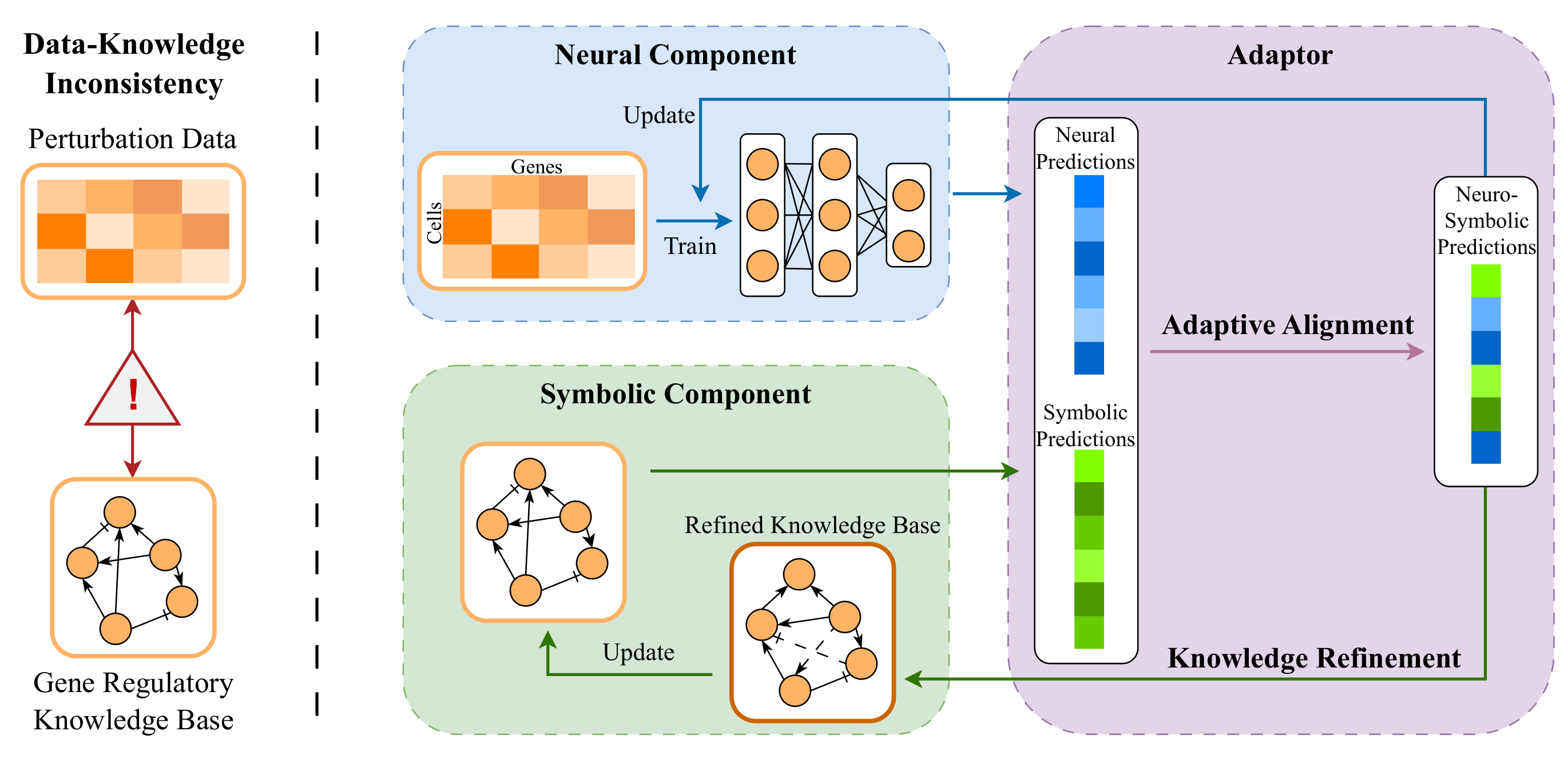
- We built a framework to solve inconsistencies between neural prediction of genetic perturbation response, driven by experimental data, and symbolic reasoning on prior knowledge of gene interactions.
- Welcome to take a look at our GitHub repo and ArXiv preprint!